
Knock-In Mice vs. Transgenic Mice:
What You Need to Know
Knock-in and transgenic mice are foundational tools in biomedical research, yet their strategic differences drive distinct experimental outcomes and value. Both play key roles and accelerated our knowledge of almost every human condition and the development of medicines and promising modalities like cell and gene therapies in the past decades [1-4]. However their design, genetic targeting precision, and applications set them apart significantly for academic and translational investigations.
Definitions and Core Differences
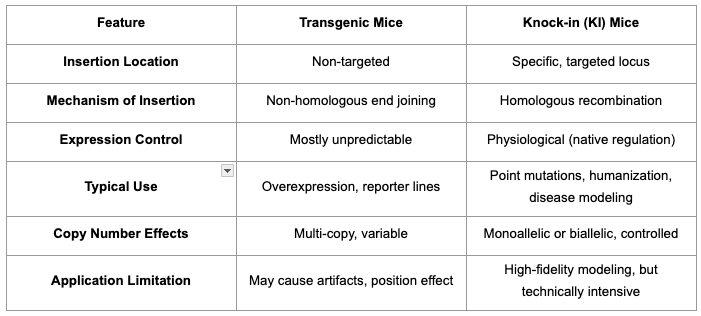
- Transgenic mice carry exogenous genetic material, introduced into the genome, typically by pronuclear microinjection or viral vectors. This integration is not targeted, which means the exogenous DNA (transgene) can land at unpredictable loci, often resulting in variable expression levels dependent on position effects, copy number, and endogenous regulatory context [2,4].
- Knock-in (KI) mice are generated by site-specific insertion of a genetic sequence (by homologous recombination) at a predefined genomic locus. Knockin constructs can replace, mutate, or augment native alleles with high fidelity, preserving endogenous regulatory environments and spatial/temporal gene expression patterns [3,4].
Scientific Utility & Experimental Considerations
- Knock-in mice excel when faithful modeling of endogenous gene function, or the introduction of precise mutations, reporters, or human alleles is critical. These models are invaluable for studying gene-disease relationships, post-transcriptional modifications, or specific humanized traits. Technologies like CRISPR and advanced homologous recombination strategies now enable the rapid development of knockin models with efficiency and accuracy previously unattainable in recent years.
- Transgenic mice remain indispensable for applications requiring overexpression, rapid proof-of-principle studies, and less stringent locus control, yielding strong, ubiquitous, or tissue-specific expression by coupling transgenes to potent promoters. However, the random nature of integration can trigger position effects, silencing, or confounding phenotypes, demanding careful validation and often multi-line analysis.
Knock-ins typically add or replace sequences at a defined locus. They can also be designed to inactivate an allele by inserting stop cassettes, frameshifts, or recombinase-dependent traps. Transgenics are not inherently unpredictable; classic random lines can vary, yet safe-harbor targeting at Rosa26 or H11 yields stable single-copy expression with fewer position effects [5].
Modern Advances and Value
In combination with modern genome engineering, knock-in and transgenic strategies now operate as a flexible toolkit that can be layered to answer mechanistic and translational questions with precision. Inducible and conditional systems such as Cre, Flp, Dre, and ligand-responsive variants like CreERT2 or Tet enable tissue, cell-type, or time-specific control. Intersectional logic using two drivers narrows expression to defined subpopulations, and lineage or activity reporters can record recombination history, cell state, and circuit function in vivo [4].
Humanization extends the reach of knock-in methods. Entire mouse exons, domains, or loci can be replaced with human orthologous sequences, preserving native regulation while enabling antibody epitope matching, small-molecule pharmacology, and biomarker development [4].
Safe-harbor or landing-pad workflows at Rosa26 or H11 provide single-copy cassette exchange with fewer position effects. This accelerates iteration of reporters, effectors, and therapeutic payloads. Multifaceted allele designs can combine knock-in cassettes with floxed elements or Lox-Stop-Lox gates to convert reporters into knockouts upon recombination. Somatic and mosaic modeling further expands scope. AAV or LNP delivery introduces editors postnatally for organ-specific perturbations and parallel testing of multiple variants within a single animal [5].
Conclusions
For scientists developing disease models, drug testing platforms, or fundamental genetic interrogation tools, knock-in mice offer unmatched precision, recapitulating native gene function and mutation effects with fidelity. Transgenic mice, while sometimes less precise, provide speed and versatility crucial for broad phenotype screening or studying gene overexpression. Both remain indispensable pillars in the genetics research toolkit, but a careful match between model type and research objective will yield the most robust, translationally relevant discoveries.
References
- Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (2002). https://doi.org/10.1038/nature01262
- Ohtsuka, M., Miura, H., Mochida, K. _et al._ One-step generation of multiple transgenic mouse lines using an improved Pronuclear Injection-based Targeted Transgenesis (i-PITT). BMC Genomics 16, 274 (2015). https://doi.org/10.1186/s12864-015-1432-5
- Mario R. Capecchi ,Altering the Genome by Homologous Recombination.Science244,1288-1292(1989). https://doi.org/10.1126/science.2660260
- Clark JF, Dinsmore CJ, Soriano P. A most formidable arsenal: genetic technologies for building a better mouse. Genes Dev. 2020 Oct 1;34(19-20):1256-1286. https://doi.org/10.1101/gad.342089.120
- Yu Z, Shi J, Zhang J, Wu Y, Shen R, Fei J. Comparative study on the expression characteristics of transgenes inserted into the Gt(ROSA)26Sor and H11 loci in mice. Acta Biochim Biophys Sin (Shanghai). 2024 May 15;56(11):1687-1698. https://doi.org/10.3724/abbs.2024081
